Solubis tools in YASARA
Contents
- 1 Preface
- 2 Aggregation Predictors
- 3 FoldX energies
- 4 Complete analysis of Object
- 5 Complete analysis of a Molecule
- 6 Mutate Residue
- 7 SOLUBIS run on complete molecule
- 8 SOLUBIS run in marked region
- 9 Save last calculation
- 10 Configure plugin
- 11 Configure plugin for FoldX
- 12 References to the FoldX methodology
Preface
In this section we explain the various SOLUBIS functionalities inside YASARA. It is recommended that you have practiced with YASARA before. But don't worry, all commands are straightforward and easy to use. That's the point of the plugin. However, we will refer to YASARA specific nomenclature such as Objects and Molecules. Their definition can be found in the YASARA documentation on your computer in yasara/doc/index.html.
The output of the SOLUBIS calculations are always printed to the YASARA console when the plugin ends. The console should open by default, but when it fails, you can enter the console by pressing the spacebar once or twice.
All FoldX tools in YASARA can be accessed by clicking:
Analyze > SOLUBIS > ...
Aggregation Predictors
The two aggregation predictors used in this SOLUBIS plugin are TANGO [1] and WALTZ [2].
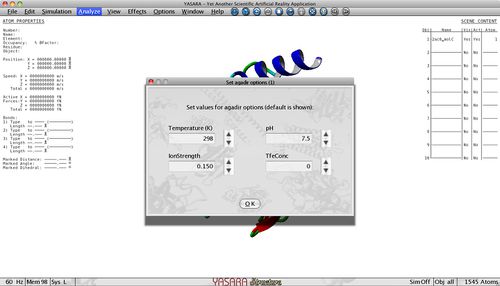
Tango predicts the aggregation propensity of an input polypeptide sequence and returns the beta-aggregating segments that have a high tendency to nucleate protein aggregation through the formation of intermolecular beta-sheets. In contrast to TANGO, WALTZ is focused on the distinction between amyloid and amorphous aggregates. Aggregation tendency is influenced by pH, temperature, ionic strenght and protein concentration so these parameters can be adjusted to fit your experiment. All these parameters are set to the default values but can be changed in the Options menu.
FoldX energies
When you perform a SOLUBIS run, you can select whether you want to calculate the effect of a mutation on protein stability. Therefor we make use of FoldX [3]. The main focus of FoldX is the prediction of free energy changes, e.g. what happens to the free energy of the protein when we mutate an Asp to a Tyr? FoldX will then calculate the free energy of the wild type (WT) and the mutant (MT) and make the difference:
<math>\Delta\Delta</math>G(change) = <math>\Delta</math>G(MT) - <math>\Delta</math>G(WT)
FoldX is trained to make <math>\Delta\Delta</math>G(change) approach experimental values.
As a rule of thumb we use:
<math>\Delta\Delta</math>G(change) > 0 : the mutation is destabilizing
<math>\Delta\Delta</math>G(change) < 0 : the mutation is stabilizing
The error margin of FoldX is approximately 0.5 kcal/mol, so changes in that range are insignificant.
Complete analysis of Object
With this command you can run one or both aggregation predictors on an object of choice. This will result in a PDB (the original or a new one) where the aggregation-prone regions in the protein are colored, if desired according to strength.
In the Option menu, you can define the specifications of an aggregating stretch:
- Window treshold: What is the minimum score for each residue to belong to an aggregating stretch. If you higher this treshold, you select only very strong aggregating stretch. On default we assume that a contiguous stretch of 5 residues with a score of at least 5 is prone to aggregation.
- Minimum window size: What is the minimal length of the aggregatin stretches you want to output.
- Flank size: Aggregating stretches are flanked by so-called gatekeepers that oppose aggregation. Here you define how many gatekeepers on each side are returned.
You can also specify the way of visualization.
In the first panel you select whether the aggregating stretches need to be indicated in the original pdb or in a new pdb (that is automatically loaded) . In the second panel, you set how you want the color the zones. If you select " All same color" , aggregating stretches predicted by TANGO or WALTZ are colored in resp. red and blue. If these aggregating stretches overlap, it is colored in magenta. If you select "Color gradient by aggregation tendency" the stretch is colored according to its strenght, ranging from yellow to red and from cyan to blue for resp TANGO and WALTZ. Weak aggregating stretches are defined as having a score below 50, strong aggregating stretches have a score above 7.
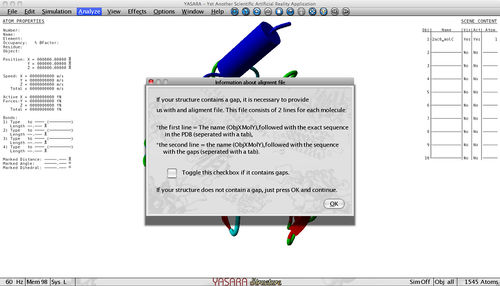
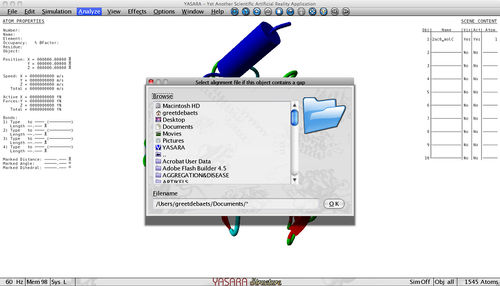
At last, the plugin also asks you whether your structure contains a gap.
As TANGO and WALTZ are sequence based predictors it is important to provide the complete sequence if your sequence contains a gap. If this is not the case, you just press ok and continue. Otherwise you provide us with an alignment file that contains 2 lines for each molecule, f.e. :
Obj1MolA\tILT---IITL Obj1MolA\tILTLLLIITL
The first line contains the name of the incomplete PDB structure (ObjXMolY), followed with a tab and the sequence as present in the PDB with '-' indicating that there is a gap at that position. The second line again contains the name of the incomplete PDB structure (ObjXMolY), followed with a tab and the complete sequence. Both sequences should have the same length and it is important that the first line contains exactly the same sequence as in the structure with the additional '-' indicating a gap.
Complete analysis of a Molecule
Mutate Residue
SOLUBIS run on complete molecule
SOLUBIS run in marked region
Save last calculation
This option lets you specify a target folder and/or filename prefix for the beginning of all files of the last calculation to be saved.
The first selection window lets you choose whether you want to save all files or just the FOLDXSUMMARY.out file. This file contains the identified aggregating stretches after a 'complete analysis of object/molecule' run or the effect of the calculated mutations after a 'mutate residue' or 'SOLUBIS' run.
In the next selection window you can either select a single folder or a folder and filename prefix:
- Folder: E.g. selecting c:\testrun\ will save all last calculation files to that folder.
- Folder with filename prefix: E.g. selecting c:\testrun\MyRun will save all last calculation files to the testrun folder and put MyRun_ before every filename. In this way, it is possible to save more than one calculation in the same folder by using different prefixes.
Configure plugin
See the section Installation and first use for explanation of this option.
Configure plugin for FoldX
Only necessary if you also want to calculate the effect on protein stability for the mutations. See the section Installation and first use for explanation of this option.
References to the FoldX methodology
- ↑ Fernandez-Escamilla, A.M., Rousseau, F., Schymkowitz, J. and Serrano, L. (2004) Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins, Nat Biotechnol, 22, 1302-1306.
- ↑ Maurer-Stroh, S., Debulpaep, M., Kuemmerer, N., Lopez de la Paz, M., Martins, I.C., Reumers, J., Morris, K.L., Copland, A., Serpell, L., Serrano, L., Schymkowitz, J.W. and Rousseau, F. (2010) Exploring the sequence determinants of amyloid structure using position-specific scoring matrices, Nat Methods, 7, 237-242.
- ↑ Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucl. Acids Res. 33:W382-8 (2005)